Abstract
Background: Mutations in the gene coding for isocitrate dehydrogenase isoform 2 (IDH2) frequently occur in acute myeloid leukemia (AML). IDH2 is an interesting target for precision medicine, as small molecule oral IDH inhibitors have been developed for specific targeting. Generally, IDH2 IDH mutations are considered early events in leukemogenesis, however, subclonal cases were also reported. Moreover, data on the presence and role of IDH2 mutations in other hematological malignancies remains scarce.
Aim: (1) Analysis of IDH2 mutation frequencies and mutation sites in 3096 patients with 28 different hematological malignancies. (2) Comparison of variant allele frequencies (VAF) of IDH2 and co-occurring mutations for differentiation between presence of IDH2 in main clone or subclone. (3) Analysis of additional cytogenetic alterations.
Methods: Whole-genome sequencing (WGS) was performed for 3096 patients (median coverage 100x). 151bp paired-end reads were generated on NovaSeq 6000 and HiSeqX instruments (Illumina, San Diego, CA). As no sample specific normal tissue was available, a so-called Tumor/Unmatched normal (TUN) workflow was used to reduce technical artefacts and germline calls. All reported p-values are two-sided and were considered significant at p<0.05.
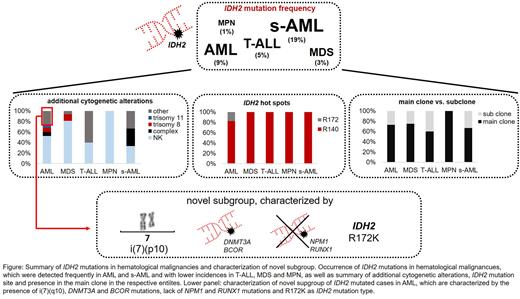
Results: In the total cohort of 3096 patients, in 91 cases (3%) a mutation in IDH2 was detected, comprising AML (63/675, 9%), MDS (16/577, 3%), T-ALL (5/101, 5%), MPN (4/306, 1%) and s-AML (3/16, 19%). All mutations were detected in the known hotspots R140 and R172, with R140Q found most frequently (n=76, 84%), followed by R172K (n=11, 12%), R140L (n=3, 3%) and R140W (n=1, 1%). Of note, R172K mutations were restricted to AML. The median VAF of IDH2 in the total cohort was 0.43 (range 0.03 - 0.56). In the subgroups, it was 0.42, 0.44, 0.36, 0.49 and 0.42 for AML, MDS, T-ALL, MPN and s-AML, respectively. The most frequent co-occurring mutations comprised SRSF2 (in 45% of IDH2 mutated cases), DNMT3A (41%), ASXL1 (29%), NPM1 (26%), STAG2 (18%) and RUNX1 (15%). Regarding co-mutations in spliceosome genes (SF3B1, SRSF2, U2AF1, ZRSR2), beside SRSF2 only U2AF1 mutations were detected recurrently (n=4), whereas mutations in SF3B1 (n=1) and ZRSR2 (n=0) were (almost) completely absent. Comparison of the IDH2 VAF with the VAF of co-mutations revealed that in 73% (66/91) of cases IDH2 was found in the main clone (AML: 73%, MDS: 75%, T-ALL: 60%, MPN: 100%, s-AML: 67%). Cytogenetic analysis of IDH2 mutated cases in the total cohort revealed a normal karyotype in 58% of cases, a trisomy 8, a trisomy 11 or a complex karyotype in 7%, 4% and 6% of cases, respectively. Interestingly, in 3 AML cases, a i(7)(p10) was observed, which constitutes a rather rare cytogenetic alteration. To investigate this further, in an unselected cohort of all cases sent to our laboratory between 08/2015 and 07/2022, a total of 34 cases with i(7)(p10) were identified. The majority of these cases comprised AMLs (n=23, 68%), but also MDS (n=4), lymphomas (n=6) and ALL (n=1) cases were observed. For 19/34 cases, a molecular genetic analysis was available, revealing an IDH2 mutation in 15/19 cases. Additionally, DNMT3A (15/19) and BCOR (7/19) mutations were frequently detected (all DNMT3A mutations co-occurred with IDH2 mutations) whereas other mutations were rather rare (SRSF2: 2/19 cases; ASXL1: 1/19; NPM1, STAG2, RUNX1: 0/19). Of note, all analyzed AML cases with i(7)(p10) harbored an IDH2 mutation. Further, an association with the hot spot R172K was found (in 18/19 cases, p<0.001).
Conclusions: (1) Mutations in IDH2 are exclusively detected in myeloid neoplasms, with the exception of T-ALL. Differences were observed between malignancies regarding the mutation site, as R172K were detected in AML only. (2) Although IDH2 mutations generally seem to be an early event in leukemogenesis, nevertheless in ~25% of cases IDH2 is only present in a subclone, questioning the use of IDH inhibitors in these cases. Hence, consideration of VAF and not only mutational state seems to be important for treatment decision. (3) A novel subgroup within IDH2 mutated cases (in particular in AML) was identified that is characterized by the presence of i(7)(p10), DNMT3A and BCOR mutations and the IDH2 mutation site R172K. NPM1 and RUNX1 mutations were not detected in this subgroup. The outcome of patients with this subgroup has to be determined in future studies (see figure).
Disclosures
Stengel:MLL Munich Leukemia Laboratory: Current Employment. Meggendorfer:MLL Munich Leukemia Laboratory: Current Employment. Walter:MLL Munich Leukemia Laboratory: Current Employment. Baer:MLL Munich Leukemia Laboratory: Current Employment. Nadarajah:MLL Munich Leukemia Laboratory: Current Employment. Hutter:MLL Munich Leukemia Laboratory: Current Employment. Kern:MLL Munich Leukemia Laboratory: Current Employment, Other: Ownership. Haferlach:Munich Leukemia Laboratory: Current Employment, Other: Part ownership. Haferlach:MLL Munich Leukemia Laboratory: Current Employment, Other: Ownership.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal